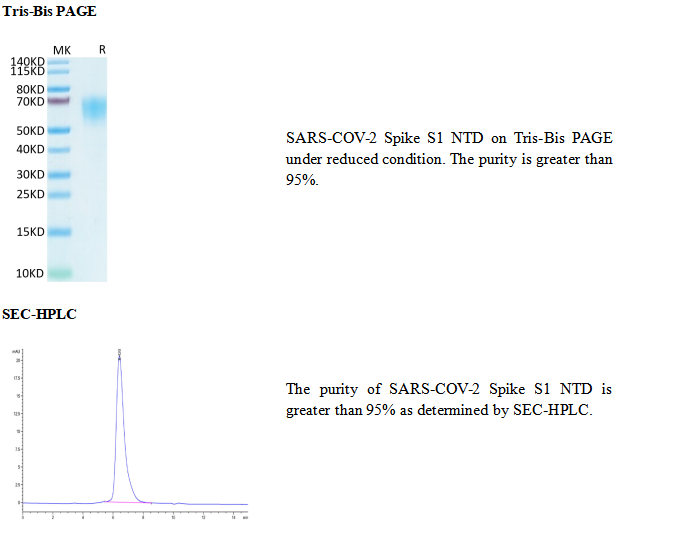
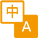SARS-CoV-2, which causes the global pandemic coronavirus disease 2019 (Covid-19), belongs to a family of viruses known as coronaviruses that also include MERS‑CoV and SARS-CoV-1. Coronaviruses are commonly comprised of four structural proteins: Spike protein (S), Envelope protein (E), Membrane protein (M) and Nucleocapsid protein (N). The SARS-CoV-2 S protein is a glycoprotein that mediates membrane fusion and viral entry. The S protein is homotrimeric, with each ~180-kDa monomer consisting of two subunits, S1 and S2. In SARS-CoV-2, as with most coronaviruses, proteolytic cleavage of the S protein into S1 and S2 subunits is required for activation. The S1 subunit is focused on attachment of the protein to the host receptor, while the S2 subunit is involved with cell fusion. The S1 subunit can be further divided into an N-terminal domain (NTD) and a receptor binding domain (RBD). The SARS-CoV-2 NTD shares 50% and 20% amino acid (aa) sequence identity with the NTD of SARS-CoV-1 and MERS-CoV, respectively. The NTD is reported to bind L-SIGN and DC-SIGN in cells that don't express the ACE-2 receptor. Despite being heavily glycosylated, the NTD is capable of eliciting an immune response to produce potent neutralization antibodies, although at a reduced level than the ones targeting the RBD. Three immunogenic regions have been identified in the NTD: aa 14-20, aa 140-158, and aa 245-264. Antibody cocktails targeting both NTD and RBD could provide better protection against SARS-CoV-2 infection.
高纯度、高活性、低内毒素、高批间一致性
产品数据
-25 ~ -15℃保存,收到货之后有效期1年。 复溶后, 无菌条件下,-85 ~ -65℃保存,3个月有效期。